|

AoE Research Programme |
||||||
 |
||||||
Fig 1
|
||||||
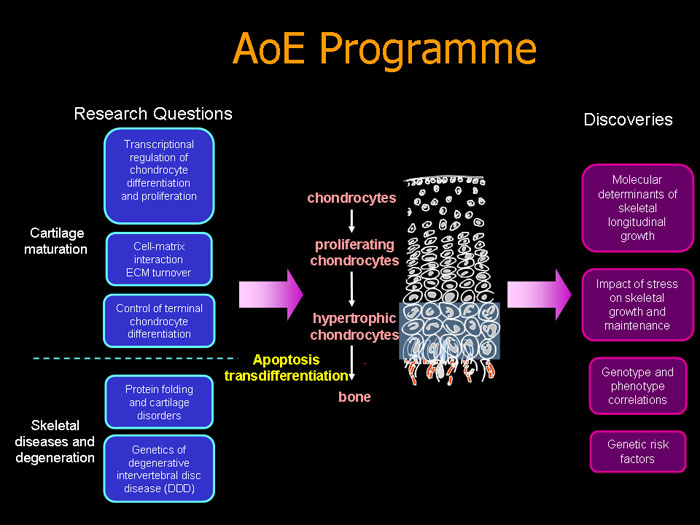
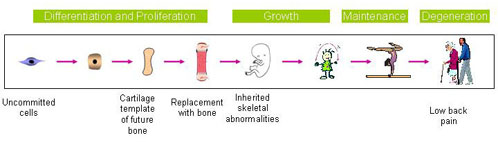
The adult skeleton, comprising over 200 elements of cartilage and bone, is formed according to a developmental programme set up by gene activities at very early stages, then maintained by continuous biosynthetic and remodelling activity throughout life. Three cell types, chondrocytes (cartilage cells), osteoblasts (bone cells) and osteoclasts (bone-resorbing cells) execute this programme to form and maintain the skeleton. Alterations in gene structure and expression can cause disorders ranging from fetal death to life-long disability, or to an early onset of skeletal degeneration. In recent years,we have gained much insight into the formation of the skeleton through studying the genetics and molecular basis of skeletal disorders in humans and mice. During the development of the mammalian skeleton, differentiation and linear growth are mediated by the growth plate cartilage located at the ends of growing bone via endochondral ossification. This is a highly coordinated programme during which chondrocytes differentiate, proliferate, mature and hypertrophy and the different stages become organized in columns in the growth plate and bone replaces calcified cartilage. This programme is controlled by a complex network of regulatory molecules and cell-extracellular matrix (ECM) interactions. Many questions remain unresolved. Which genes, proteins and pathways determine the columnar organization of the growth plate? How does this control longitudinal growth of the skeleton, cartilage and bone mineralization and bone mass in post-natal life? How do different forms of stress, such as misfolded proteins, affect skeletal growth and maintenance and contribute to skeletal disorders? What are the genetic predisposing factors and molecular pathogenetic basis of common degenerative diseases of the skeleton? The powerful tools now available ― tailored genetically modified organisms, coloured proteins, large-scale gene and protein expression profiling and bioinformatics make all of these questions accessible to investigation. As summarized in Fig. 1, this AoE programme focuses on key issues at the frontier of developmental biology and skeletal research related to the formation and growth of cartilage and bone and the maintenance of skeletal function . Through genetics and functional genomics we aim to understand: A. Genes, pathways and networks regulating cartilage formation and maturation in skeletal growth, through studying
B. Skeletal disorders and degeneration, focusing on
|
||||||
Research Strategy A major experimental strategy in the AoE programme is to use tissue- and stage-specific and systematic ablation of genes and pathways in mouse models to understand genes and their functions and how distinct signalling pathways coordinate the proper cascade of differentiation within the mammalian growth plate in the fetal and post-natal period. We are also using simple systems such as the nematode C. elegans for basic gene function studies and hypotheses testing. The genes of C. elegans operate in a simpler cellular architecture than vertebrate genes. C. elegans does not have a skeleton, but it does possess every signal transduction pathway found in mammalian systems and homologues of human genes perform analogous functions. We are also using large-scale pairwise comparisons of expression profiles between normal and mutant states and among different developmental stages, using DNA microarrays and proteomics as part of our strategy to characterize the molecular pathways involved in the formation of the growth plate. These technologies combined with bioinformatics will allow us to increase the scale and power of analysis by several orders of magnitude compared with current subtractive/differential library screening techniques. |
||||||
 |
||||||
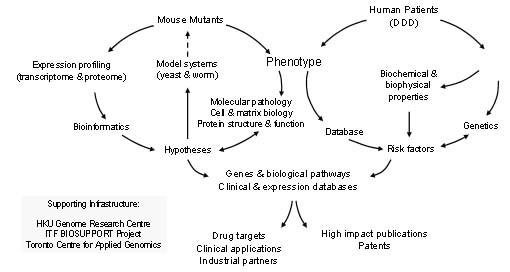
By correlating the molecular mechanisms and genetic factors revealed in the Genetics studies with knowledge gained from the loss of gene function experiments and the biochemical studies carried out by the protein Science Team we aim to obtain fundamental insight into skeletal development and growth and the molecular pathogenesis and genetic influences governing skeletal disorders and degeneration. Through this integrated research programme, we will determine the combination of factors that is necessary to maintain cartilage phenotypes. Such information is extremely important for the development of cell and tissue-based therapies for genetic and acquired skeletal disorders that require expanding specific populations of differentiated chondrocytes in vitro. The answers we provide will be applicable beyond DDD to major diseases of skeletal tissues such as osteoarthritis and osteoporosis.
|
||||||
Research organisation The research of the AoE programme is facilitated through the establishment of
Research Teams Three expertise teams have been set up: the Functional Genomics team, Protein Science team and Genetics team. Establishing these teams facilitates regular frequent research meetings and interactions amongst AoE Members in smaller groups.
|
||||||
 |
||||||
The group meeting coordinators for the groups are:
|
||||||