Academic Staff

Professor QIAN, Chengmin 錢程民
- BSc (Anhui Normal U); PhD (Nanjing U)
- Associate Professor
- Cell and Cancer Biology
- Epigenetics
- DNA Damage Response
Epigenetic changes play essential roles in normal development and human diseases. Aberrant epigenetic alternations have been implicated in many human cancers and inherited syndromes. To understand how epigenetic modifications occur during normal development, adult cell renewal and disease will significantly help to prevent, diagnose and treat epigenetic-related diseases. A major challenge in the field of epigenetics is to define the connection between distinct epigenetic modification and specific biological function. My current research is directed at understanding molecular mechanisms of epigenetic modifications at atomic resolution by using protein NMR and X-ray crystallography. We are studying chromatin modifying enzymes that establish different epigenetic modifications, and readout proteins that interpret epigenetic modifications. Our ultimate goal is to apply structure-based rational design method to develop small chemical compounds capable of modulating these epigenetic processes under physiological conditions.
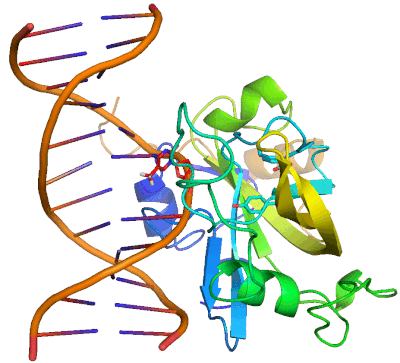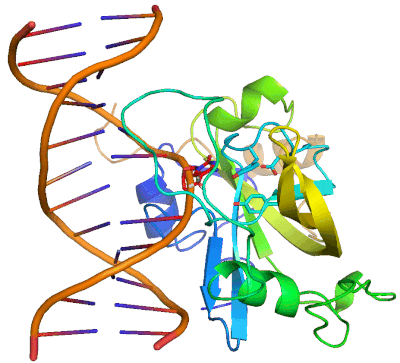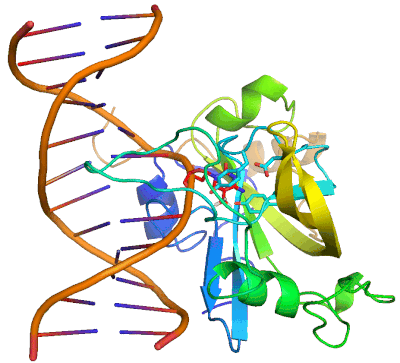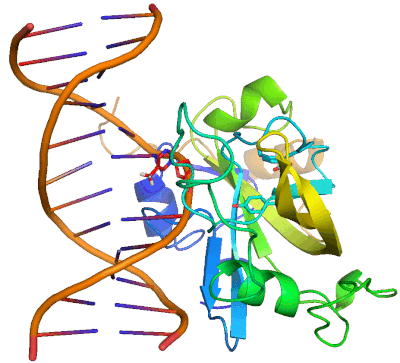
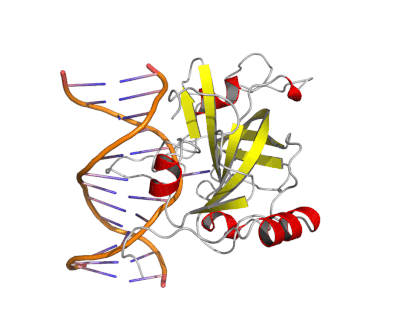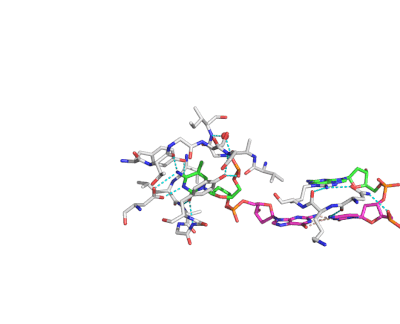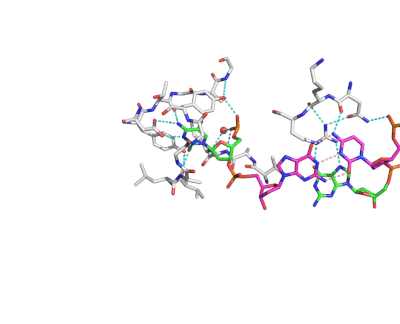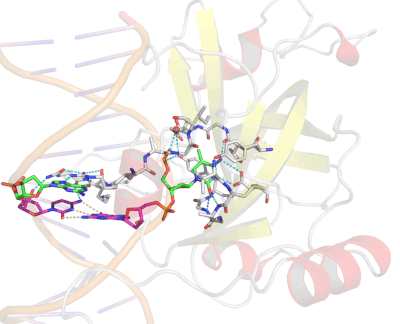
- Structure and Mechanism of Protein Module in Epigenetics
- Structural Biology of DNA Damage & Repair
- We currently have the opportunity to recruit a highly-motivated postdoctoral fellow to work on the structural mechanisms of intra-S-phase checkpoint activation in response to DNA damage. The candidate should have a background in protein crystallography or protein NMR.
- A research assistant position is available in the lab. The candidate is required to have hands-on experience with protein cloning, expression and purification, a background in structural biology is preferred. The successful candidate will participate in our ongoing studies on structure & mechanism of novel tumor suppressors involved in damaged DNA repair. Applicants should email their CV and a summary of research experience to Dr. Chengmin Qian (cmqian@hku.hk)
- Du YM, Yan YX, Xie S, Huang H, Wang X, Ng R, Zhou MM, Qian C*. (2021) Structural mechanism of bivalent histone H3K4me3K9me3 recognition by the Spindlin1/C11orf84 complex in rRNA transcription activation. Nature Communications. 12:949. PMID: 33574238. [Download pdf]
- Xie S, Qian C*. (2018) The growing complexity of UHRF1-mediated maintenance DNA methylation. Genes. 9(12):600. PMID: 30513966. (Invited review for special issue of “Role of DNA Methyltransferases in the Epigenome”.)
- Li T, Wang LS, Du YM, Xie S, Yang X, Lian FM, Zhou ZJ, Qian C*. (2018) Structural and mechanistic insights into UHRF1-mediated DNA methylation maintenance. Nucleic Acids Research. 2018. Feb 19. 46(6):3218-3231. [Download pdf]
- Yan L, Xie S, Du YM, Qian C*. (2017) Structural insights into BAF155 and BAF47 complex formation. Journal of Molecular Biology. 429(11):1650-1660. [Download pdf]
- Lian FM, Xie S, Qian C*. (2016) Crystal structure and SUMO binding of Slx1-Slx4 complex. Scientific Reports. 6:19331.
- Aguilo F, Li S, Balasubramaniyan N, Sancho A, Benko S, Zhang F, Vashisht A, Rengasamy M, Andino B, Chen C, Zhou F, Qian C, Zhou MM, Wohlschlegel J, Hzng W, Suchy F, Walsh MJ. (2016) Deposition of 5-Methylcytosone on enhancer RNAs enables the coactivator function of PGC-1α. Cell Reports. 14(3): 479-492.
- Xie S, Mortusewicz O, Ma HT, Herr P, Poon RY, Helleday T, Qian C*. (2015) Timeless interacts with PARP-1 to promote homologous recombination repair. Molecular Cell. 60(1): 163-176. (Recommended by Faculty of 1000: Rated as Exceptional) [Download pdf]
- Lai, Chang YY, Hu L, Yang Y, Chao A, Du ZY, Tanner JA, Chye ML, Qian C, Ng KM, Li H, Sun H. (2015) Rapid labeling of intracellular His-tagged proteins in living cells. Proc Natl Acad Sci U S A. 112(10): 2948-2953.
- Xie S, Lu Y, Jakoncic J, Sun H, Xia J, Qian C*. (2014) Structure of RPA32 bound to N-terminus of SMARCAL1 redefines the binding interface between RPA32 and its interacting proteins. FEBS Journal. 281(15): 3382-3396.
- Xie S, Jakoncic J, Qian C*. (2012). UHRF1 Double Tudor Domain and the Adjacent PHD Finger Act Together to Recognize K9me3-containing Histone H3 Tail. Journal of Molecular Biology 415, 318-28.
- Qian C, Li S, Jakoncic J, Zeng L, Walsh M, Zhou M (2008). Structure and hemimethylated CpG binding of the SRA domain from human UHRF1. Journal of Biological Chemistry 283, 34490-4.
- Qian C, & Zhou, M.-M. (2006). SET domain protein lysine methyltransferase: Structure, specificity and catalysis. Cellular and molecular life sciences. 63: 2755-2763.
- Qian C, Wang X, Manzur K, Sachchidanand, Zeng L, Farooq A, Zhou M (2006). Structural insights of specificity and catalysis of a dimeric viral SET domain, a histone H3 lysine-27 methyltransferase. Journal of Molecular Biology 359, 86-96.
- Qian C, Zhang Q, Li S, Zeng L, Walsh M, Zhou M (2005). Structure and chromosomal binding of SWIRM domain. Nature Structural and Molecular Biology 12, 1078-1085. (Recommended by Faculty of 1000)
- Research Grants Council (RGC) - General Research Fund (GRF)
- 2018-2021 “Structural and mechanistic insights into Timeless in DNA damage response”
- 2017-2020 “Identification and characterization a novel protein complex in rRNA transcription”
- 2016-2019 “New Mechanistic Insights into DNMT1-mediated Maintenance DNA Methylation”
- 2015-2018 “Structural & Functional Analysis of BAF Chromatin Remodeling Complex”
- 2014-2017 “Structural and Mechanistic Studies of Human SLX4 Complex in Interstrand Crosslink Repair”
- 2013-2016 “Structure-Based Mechanistic Insights into Timeless-Involved Replication Fork Protection”
- 2012-2015 “Structural and Mechanistic Characterization of XPA-associated Complexes in Nucleotide Excision Repair Pathway”
- 2011-2014 “Structural and Mechanistic Analysis of Human SMARCAL1”
- 2010-2013 “Structural and Functional Analysis of Oncoprotein UHRF1”
Last update: 2021-03-03
