Academic Staff

Professor TANNER, Julian Alexander 唐柱霖
- BSc (Bristol); PhD (Imperial College London)
- Dexter H C Man Family Professor in Medical Science
- Director of the Common Core, The University of Hong Kong
- Associate Dean (Health Sciences Education), Li Ka Shing Faculty of Medicine
- Nucleic Acid Engineering
- Chemical Biology
- Student Learning
Professor Julian A. Tanner is Director of the Common Core and Associate Dean (Health Sciences Education) of the HKUMed T&L Deanery.
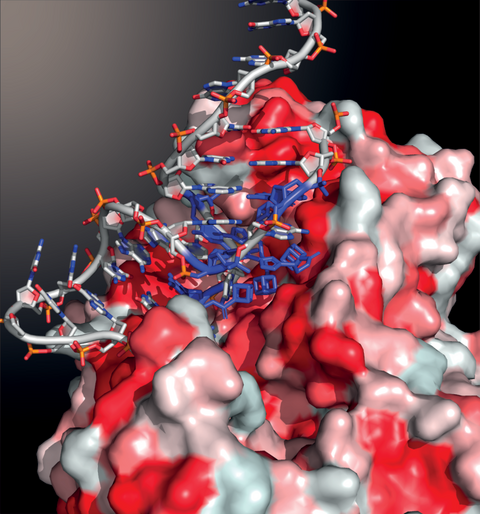
Evolution of abiotic cubane chemistries in a nucleic acid aptamer allows selective recognition of a malaria biomarker as published in PNAS in July 2020 as a collaboration between the University of Hong Kong and Pasteur Institut, Paris. The image shows a cubane-modified aptamer in blue which was evolved in our laboratory to specifically recognise a malaria protein biomarker depicted by the red and white surface.
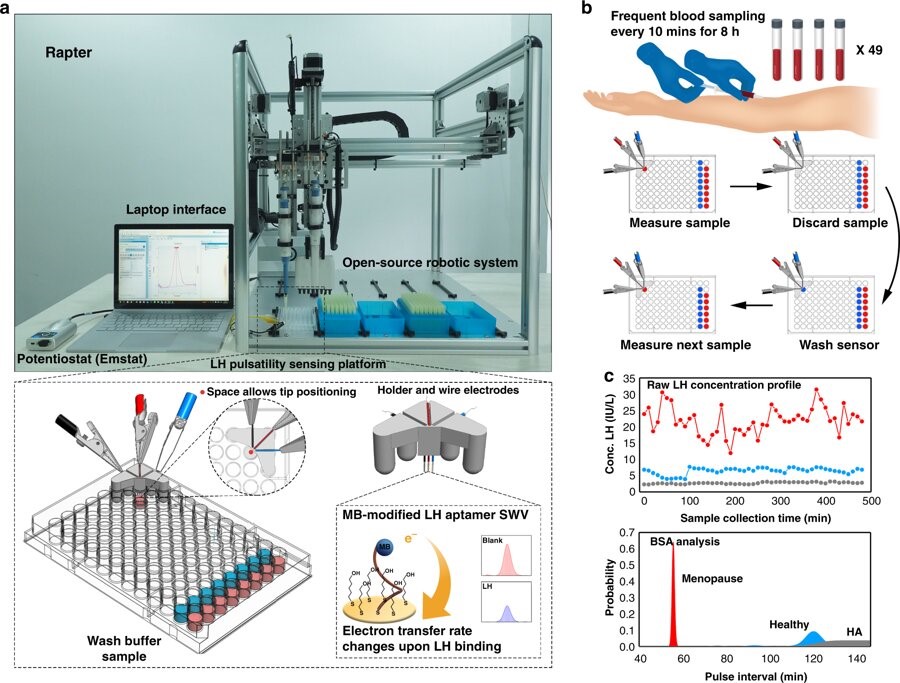
Robotic APTamer-enabled electrochemical reader (RAPTER) for luteinising hormone sensing to diagnose reproductive disorders as published in Nature Communications in Feb 2019 as a collaboration between the University of Hong Kong and Imperial College London.
-
Julian's research field is chemical biology, with a particular focus on directed nucleic acid evolution for translational biomedical application. The Tanner lab brings together interdisciplinary experimental approaches at the interface of biomedical science, engineering and chemistry to tackle major medical challenges. His highly collaborative research team is developing new technologies for point-of-care biosensing, diagnostics and therapeutics in a variety of contexts, including for infectious disease, infertility, various cancers and sepsis. The team is also active into research into nucleic acid structure and function, investigating the interface of aptamers and DNA nanotechnology through interdisciplinary apporoaches. The team publishes frequently in leading journals including recently in Nature Communications, PNAS, Angewandte Chemie, Biosensors & Bioelectronics, Small, and holds over a dozen patents.
-
Julian also plays major roles in T&L at HKU. He is Director of the Common Core across the University of Hong Kong, and is Associate Dean (Health Sciences Education) of the HKUMed T&L Deanery. He is the former Director of the BBiomedSc degree programme (2018-2024), and played a major role in reform of the first two years of the MBBS curriculum in a former Assistant Dean role at HKUMed (2018-2024). He is co-author of one of the leading textbooks of Chemical Biology – Essential of Chemical Biology (Wiley), with a second edition recently published in early 2024. Julian has particular interest in how to build the best transdisciplinary environment for student achievement at the nexus between T&L, research and innovation for students across all disciplines and faculties of the University.

- Kinghorn, A.B., Guo, W., Wang, L., Tang, M.Y.H., Wang, F., Shiu, S.C.C., Lau, K.K., Jinata, C., Dey Poonam, A., Shum, H.C.* & Tanner, J.A.* Evolution Driven Microscale Combinatorial Chemistry in Intracellular Mimicking Droplets to Engineer Thermostable RNA for Cellular Imaging, Small, 2409911 (2025)
-
Lo, Y., Siu, R.H.P., Tran, C., Jesky, A.B., Kinghorn, A.B. & Tanner, J.A. An aptamer/CRISPR electrochemical (ACE) biosensor for Plasmodium falciparum histidine-rich protein II and SARS-CoV-2 nucleocapsid protein, Microchem J, 212, 113176 (2025)
-
Siu, R.H.P., Jesky, R.G. Fan, Y.J., Au-Yeung, C.C.H., Kinghorn, A.B., Chan, K.H., Hung, I.F.N. & Tanner, J.A. Aptamer-Mediated Electrochemical Detection of SARS-CoV-2 Nucleocapsid Protein in Saliva, Biosensors, 14, 10, 471 (2024)
- Whitehouse, W.L., Lo, L.H.Y., Kinghorn, A.B., Shiu, S.C.C. and Tanner, J.A. Structure-switching electrochemical aptasensor for rapid, reagentless and single-step nanomolar detection of C-reactive protein. ACS Appl Bio Mater, 7, 6, 3721-3730 (2024)
- Niogret, G., Bouvier-Muller, A., Figazzolo, C., Joyce, J.M., Bonhomme, F., England, P., Mayboroda, O., Pellarin, R., Gasser, G., Tucker, J.H.R., Tanner, J.A., Savage, G.P. & Hollenstein, M. Interrogating aptamer chemical space through modified nucleotide substitution facilitated by enzymatic DNA synthesis. Chembiochem, e202300539 (2024)
- Bhuyan, S.K., Wang, L., Jinata, C., Kinghorn, A.B., Liu, M., He, W., Sharma, R. & Tanner, J.A. Directed evolution of a G-quadruplex peroxidase DNAzyme and application in proteomic DNAzyme-aptamer proximity labeling. J. Am. Chem. Soc., 145, 23, 12726-12736 (2023)
- Guo, W.*, Kinghorn, A.B.*, Zhang, Y., Li, Q., Poonam, A.D., Tanner, J.A.* & Shum, H.C.* Non-associative phase separation in an evaporating droplet as a model for prebiotic compartmentalization. Nature Commun, 12, 1-13 (2021)
- Minopoli, A., Della Ventura, B., Lenyk, B., Gentile, F., Tanner, J. A. Offenhausser, A., Mayer, D. & Velotta, R. Ultrasensitive antibody-aptamer plasmonic biosensor for malaria biomarker detection in whole blood. Nature Commun, 11, 6134 (2020)
- Cheung, Y.W.*, Rothlisberger, P.*, Mechaly, A.E., Weber, P.P.W., Levi-Acobas, F., Lo, Y.,, Wong, A.W.C., Kinghorn, A.B., Haouz, A., Savage, G.P., Hollenstein, M.* & Tanner, J.A.* Evolution of abiotic cubane chemistries in a nucleic acid aptamer allows selective recognition of a malaria biomarker. Proc Natl Acad Sci USA, 117, 16790 (2020)
- Liang, S., Kinghorn, A.B., Voliotis, M., Prague, J.K., Veldhuis, J.D., Tsaneva-Atanasova, K., McArdle, C.A., Li, R.H.W., Cass, A.E.G.*, Dhillo, W.S.* & Tanner, J.A.* Measuring luteinising hormone pulsatility with a robotic aptamer-enabled electrochemical reader. Nature Commun, 10, 852 (2019)
- Cheung, Y.W., Kwok, J., Law, A.W.L., Watt, R.M., Kotaka, M. & Tanner, J.A. Structural basis for discriminatory recognition of Plasmodium lactate dehydrogenase by a DNA aptamer. Proc Natl Acad Sci USA, 110(40), 15967-15972 (2013)
- Miller, A.D. & Tanner, J.A. Essentials of Chemical Biology: Structure and Dynamics of Biological Macromolecules – Second Edition. John Wiley & Sons, Ltd, Chichester, UK. 560 pages (2024) (authored book)
- Khong, M.L. & Tanner, J.A. Surface and deep learning: a blended learning approach in preclinical years of medical school. BMC Med Educ, 24(1), 1029 (2024)
- Khong, M.L. & Tanner, J.A. A collaborative two-stage examination in biomedical sciences: positive impact on feedback and peer collaboration. Biochem Mol Biol Edu, 49(1), 69-79 (2021)
- Tanner, J.A., Kinghorn, A.B. & Cheung, Y.W. Aptamers. MDPI, Basel, Switzerland. 289 pages (2018) (edited book)
- Bevan, S.J., Chan, C.W.L. & Tanner, J.A. Diverse assessment and active student engagement sustain deep learning: a comparative study of outcomes in two parallel introductory biochemistry courses. Biochem Mol Biol Edu, 42(6), 474-479 (2014)
- Miller, A.D. & Tanner, J.A. Essentials of Chemical Biology: Structure and Dynamics of Biological Macromolecules. John Wiley & Sons, Ltd, Chichester, UK. 573 pages (2008) (authored book)



- ScienceDaily: New robotic sensor technology can diagnose reproductive health problems in real-time
- Phys.org: New robotic sensor technology can diagnose reproductive health problems in real-time
- 蘋果新聞: 港英研發 20元測女性不孕 微極電線檢測黃體生成素 免重複抽血
- 星島日報: 港大與英研發 快速診斷不孕症
- 東方日報: 新「單鏈DNA序列」 助快速診斷不孕症
- 信報: 云爾錄 :港大快速驗女性不孕僅需20元
- 晴報: 港大研檢測平台 速驗不孕不育
- 都市日報: 速測不孕疾病 診斷成本約廿蚊
- 經濟日報:生殖障礙難診斷 港大與倫敦帝國學院開發自動檢測平台
- 東網: 港大研不孕症快速檢測 費用僅20元
- HK01: 港大與倫敦帝國學院研傳感檢測技術快速診斷不孕不育 成本減九成
- 巴士的報: 港大與英學府開發檢測平台 查女性不孕費用僅傳統方法0.2%
- Advanced Science News: Proteins feel the pinch with DNA nanotweezers
- Burnet Institute: Burnet Institute and University of Hong Kong in new collaboration
- South China Morning Post: HKU team discovers DNA technique to detect malaria
- Medical Tribune: DNA aptamer to transform malaria diagnosis
- Aptamer: Molecular Insight and Translational Theranostics (HKU Co-PI, UGC TBRS 2021)
- Advanced Biomedical Instrumentation Centre (ABIC) InnoHK Co-PI (2021-2025)
- Toroidal DNA nanostructures for rotary catalysis (CERG GRF 2024)
- Kepler-Poinsot DNA nanostructures as sharp next-generation drug delivery vehicles (CERG GRF 2021)
- COVID-19 Point-of-Care Diagnostics in Saliva: an Aptamer-Mediated Approach (ITF COVID-19 2021)
- Da Vinci-Inspired DNA nanostructures targeted with aptamers for cancer therapeutics (CERG GRF 2020)
- Nucleic acid aptamer decorated peptide amphiphile nanofibers for regenerative medicine applications (CERG GRF 2018)
- Tanner, J.A. & Shiu, S.C.C. Facet-based nucleic acid platonic, Kepler-Poinsot polyhedral and four-dimensional tesseract for therapeutic, diagnostic and analytical applications. PCT International Application, USPTO 63/638955 (2024).
- Tanner, J.A. & Shiu, S.C.C. Nucleic acid mazzocchio and methods of making and use thereof. US20220168429A1 (2022), CH114072505 (2023), EP3953475A4 (2022)
- Tanner, J.A. & Cheung, Y.W. Sandwich and species-specific nucleic acid aptamers against Plasmodium lactate dehydrogenase for malaria diagnosis. PCT/CN2017/115895 (2017)
- Kinghorn, A.D. & Tanner, J.A. DNA display and methods thereof. WO2016134521 A1 (2015)
- Tanner, J.A., Cheung, Y.W. & Kotaka, M., Nucleic acid aptamers against Plasmodium lactate dehydrogenase and histidine-rich protein II and uses thereof for malaria diagnosis. US Patent No. 9000137, CN104245957A, EP2812453A1 (2013)
- Tanner, J.A. & Cheung, Y.W. DNA aptamers against plasmodium lactate dehydrogenase and histidine-rich protein II for malaria diagnosis and therapy. US Provisional Patent, 61/596,774 (2012)
- Tanner, J. A., Shum, K.T. & Chan, C.S.L., High-affinity nucleic acid aptamers against sclerostin protein. US2011/0294872 A1, EP2576786A1 (2011-2014)
- HKU Teaching Innovation Award (Team) (2023)
- QS Reimagine Education Silver and Bronze Awards (2023)
- Faculty Outstanding Research Output Award (2020)
- Hong Kong UGC Teaching Award (Team) (2019)
- HKU Outstanding Teacher Award (Team) (2018)
- HKU Outstanding Research Student Supervisor Award (2017)
- HKU Outstanding Young Researcher Award (2016)
- HKU Outstanding Teacher Award (Individual) (2015)
- Fellow of the Royal Society of Chemistry (FRSC) (2014)
- Dr Andrew Kinghorn - Research Assistant Professor
- Dr Simon Chi-Chin Shiu - Post-Doctoral Fellow
- Dr Louisa Lo - Post-Doctoral Fellow (Advanced Biomedical Instrumentation Centre)
- Dr William Whitehouse - Post-Doctoral Fellow (Advanced Biomedical Instrumentation Centre)
- Dr Soubhagya Bhuyan - Post-Doctoral Fellow (Advanced Biomedical Instrumentation Centre)
- Dr Lin Wang - Post-Doctoral Fellow (Advanced Biomedical Instrumentation Centre)
- Jingyu Cui - PhD student
- Weisi He - PhD student
- Amro Samir - PhD student
- Ryan Siu - PhD student
- Xueyan Wang - PhD student
- Yinuo Xie - PhD student
- Xiaotian Zhang - PhD student
- Melody Lin Wing Tung - MRes[Med] student
- Zach Smith - Technician
* I am open to enquiries from prospective PhD students and post-doctoral fellows, please contact me. Financially supported positions may be available for excellent applicants, either local or international. Hong Kong PhD Fellowship Scheme applicants are particularly welcome.
As primary supervisor:
- Dr Soubhagya Bhuyan - PhD
- Dr Cecilia Chan - PhD
- Dr Celine Chan - PhD
- Dr Yee-Wai Cheung - PhD
- Dr Mei Choi - PhD
- Dr Roderick Dirkzwager - PhD
- Dr Lewis Fraser - PhD
- Dr Chandra Jinata - PhD
- Dr Mei Li Khong - PhD
- Dr Andrew Kinghorn - PhD
- Dr Shaolin Liang - PhD (joint HKU-Imperial)
- Dr Mengping Liu - PhD
- Dr Young Lo - PhD
- Dr Eric Lui - PhD
- Dr Simon Shiu - PhD
- Dr Kato Shum - PhD
- Dr Wesley Tucker - PhD
- Dr Lin Wang - PhD
- Dr Yuanyuan Yu - PhD
- Carl Ao - MPhil
- Mari Kimura - MPhil
- Bob Lee - MPhil
- Marco Tang - MPhil
- Alvin Wai Chung Wong - MMedSc
Last update: 2025-08-21

