Sep 21, 2020
Press Release: HKUMed researchers discover a novel mechanism of multicellular pattern formation providing new insights into development
Press Release (2020-09-21):
HKUMed researchers discover a novel mechanism of multicellular pattern formation providing new insights into development
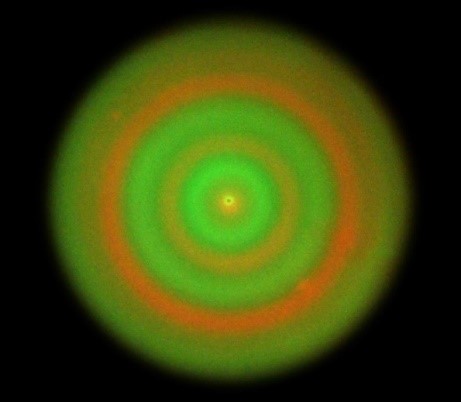
HKUMed synthetic biologists program bacteria to form multicellular biological patterns.
Living organisms exhibit amazing patterns or structures during development, such as stratified cell layers during early embryogenesis, animal skin patterns composed of different pigment cells, etc. Understanding how these patterns or structures are formed remains a fundamental scientific question. Researchers at the School of Biomedical Sciences, LKS Faculty of Medicine, The University of Hong Kong (HKUMed) collaborated with researchers at Université Paris Diderot, France and the Institute of Synthetic Biology of Chinese Academy of Sciences - Shenzhen Institutes of Advanced Technology, to conduct a study of pattern formation in artificial multicellular systems. Combining experimental biology and theoretical physics, the study identified the reciprocal control of motility to be a general mechanism for biological pattern formation and self-organization of different cell types, providing new insights into development. The findings are now published in Nature Physics [link to the publication].
Background
Pattern formation, as one of the most remarkable features of life, describes the reliable and recurrent generation of orderly structures or patterns. As early as 1952, Alan Turing, known as the father of modern computer science, proposed the “reaction-diffusion“ model to explain those naturally occurring biological patterns, which opened up a long studying process of this topic. The most crucial feature of pattern formation is its self-organization, by which biological systems can develop spatial patterns autonomously without any pre-existing positional information. At the cellular and population level, it mounts to how different cell types assume specified distribution in space and time through cell differentiation, proliferation, and migration. Studying pattern formation and clarifying their regulatory mechanisms are crucial to the understanding of important biological processes such as embryogenesis and morphogenesis. However, in most biological systems, the overwhelming complexity makes elucidating any underlying pattern forming process a challenge in the absence of any guiding principles.
Research findings
Instead of natural living systems, the research team worked with artificial living systems which are built by synthetic biology. Using E. coli as a model organism, they fabricated signaling pathways between two types of cells,which allow them to mutually control their motility in response to the local cell density of their counterpart population. When the two cell populations are programmed to enhance the motility of each other and seeded as a cell mixture on a semisolid agar plate, the initially well-mixed cell populations tend to segregate and form an orderly stripe pattern in which two cell types alternatively appeared as concentric rings. By re-designing the artificial signaling pathways, the researchers also programmed two cell populations to suppress the motility of each other. Interestingly, the mixture also autonomously developed periodic rings, however, this time, the two cell types tend to always colocalized in the rings. The research team further established a theoretical model to decipher the mechanisms of the observed pattern formation processes. Simulation of the model produced similar ring patterns that both qualitatively and quantitatively agreed with the experiments.
The development of a mature organism from a single cell is a complicated but orderly process. During the process, diverse cell types are differentiated and coordinate to form regular body layouts spatiotemporally. For example,in zebrafish, different types of pigment cells are accurately aligned to form colored stripes. In past studies, several mechanisms have been proven to play important roles in self-organization of biological systems,such as cell-cell adhesion. “Our study shows reciprocal control of motility to be a novel self-organization pathway and a general pattern formation mechanism in multicellular systems. This pattern forming mechanism explains how different cell populations could spontaneously colocalize or segregate in space and time without the guidance of any predetermined positional information or man-made interference, which is a prerequisite of the formation of more complex biological patterns and structures This finding may inspire us to check if such mechanism is at play in the pattern formation of motile cells in nature. From an engineering view, it could provide a guiding principle for tissue engineering and regenerative medicine.” remarked by Professor Jian-Dong Huang, L & T Charitable Foundation Professor in Biomedical Sciences, School of Biomedical Sciences, HKUMed, who led the research.
About the Research Team
The study was led by both HKUMed and Université Paris Diderot, France. The two lead investigators are Prof Jian-Dong Huang, L & T Charitable Foundation Professor in Biomedical Sciences, School of Biomedical Sciences, HKUMed and Dr. Julien Tailleur, CNRS Researcher, Laboratoire MSC, Université Paris Diderot. Professor Chenli Liu, the Institute of Synthetic Biology of Chinese Academy of Sciences - Shenzhen Institutes of Advanced Technology and Dr. Adrian Daerr, Université Paris Diderot participated in the research. Dr. Agnese I. Curatolo, Université Paris Diderot, Dr. Nan Zhou and Dr. Yongfeng Zhao, School of Biomedical Sciences, HKUMed, are co-first authors.
This work was supported by ANR / RGC Joint Research Scheme (A-HKU712/14), the Shenzhen Peacock Team Project, Shenzhen Science and Technology Innovation Committee Basic Science Research Grant.
Media enquiries
School of Biomedical Sciences, HKUMed
Professor Huang Jian-dong (Tel: 3917 6810 | Email: jdhuang@hku.hk)
School of Biomedical Sciences (General Office), HKUMed
Christina Chan (Tel: 3917 6334 | Email: slcchan@hku.hk)
港大醫學院發現多細胞系統生物圖案形成的全新機制 為理解生物發育提供新的見解

港大醫學院合成生物學家編碼細菌產生多細胞生物圖案。
生物體在發育過程中通常會形成多種多樣的圖案或結構,如胚胎早期發育中出現的有層次的細胞層,由不同色素細胞組成的動物皮膚花紋等。理解這些生物圖案或結構是如何形成的,長久以來是生物學最基本的科學問題之一。香港大學李嘉誠醫學院(港大醫學院)生物醫學學院與巴黎第七大學、中國科學院深圳先進技術研究院合成生物學研究所的科研團隊合作,在人工構建的多細胞生物體系中開展生物圖案形成機制的研究。此項研究緊密結合生物實驗和物理理論,揭示了多細胞群體通過相互調控運動產生自組織的基本原理及生物圖案形成的一般規律,為理解生物發育提供了新的見解。相關研究成果已刊登於科學期刊《自然-物理》。 (按此瀏覽期刊文章)
研究背景
生物圖案的形成是生命最顯著的特徵之一,也是反覆生成有序的結構或圖案。早在1952年,計算機科學之父阿蘭圖靈就提出了「反應-擴散」模型,用於解釋自然界中自發產生的生物圖案,開啓了生物圖案研究的漫長之旅。生物圖案形成的一個最重要特徵是其自組織性,即通常不需要外來干預即可自發形成。在細胞和群體水平,則體現在不同細胞群體經過分化、生長和運動等過程,形成在時空的精確排佈。研究生物圖案或結構的形成,明確其調控機制,對於理解胚胎發生和形態發生等生命過程具有重要意義。然而,自然界生物系統的多樣性和複雜性為闡明生物圖案形成背後的原理帶來了極大的挑戰。
研究發現
研究團隊轉而利用合成生物學手段來構建人工生物系統,而非研究自然存在的生物體。利用大腸桿菌作爲模式生物,研究團隊在兩群细胞間設計了人工的信號通路,使得它們通過感受對方的局部密度調控彼此的運動能力。將兩種互相增强對方運動的細胞混合並點種於半固體培養基後,原本均匀混合的兩種細胞群體趨向於分離,並形成規律的條纹圖案,在這個圖案中兩種細胞以同心圓的形式交替出現。研究團隊隨後通過重新設計信號通路,進而使得兩種細胞群體在感知對方密度後互相抑制對方的運動。有趣的是,混合細胞群體在培養基中也形成了類似的同心圆圖案,然而在該模式下,兩種細胞群體則始终聚集在一起。爲了解釋上述圖案形成背後的原理,研究團隊建立了理論模型進行研究電腦模擬產生的圖案。在定性和定量上幾乎與實驗結果一致。
生物從單細胞發育為一個成熟的個體是一個複雜但有序的過程。這一過程涉及到細胞分化為不同類型的細胞,並在空間中通過相互協調形成有序的身體結構。例如,在斑馬魚中,不同類型的色素細胞精確排佈形成彩色皮肤條紋。領導此研究的港大醫學院生物醫學學院教授、慧賢慈善基金教授(生物醫學)黃建東教授表示:「我們的研究證明了運動行爲的相互調控是一種新的自組織方式和多細胞生物體系形成圖案的一般規律。這一機制可以解釋不同的細胞群體如何在沒有外界預先存在的信號指導或人爲干預下,自發的聚集和分離。這是形成更複雜生物圖案或身體結構的先決條件。這一發現可以啓發我們去探索自然界中運動細胞的生物圖案形成是否存在類似的情況。從工程的角度來説,這也能爲今後的組織工程和再生醫學提供指導。」
關於研究團隊
本研究由港大醫學院及巴黎第七大學共同領導。兩位首席研究員為港大醫學院生物醫學學院教授、慧賢慈善基金教授(生物醫學)黃建東教授及巴黎第七大學的Julien Tailleur博士。中國科學院深圳先進技術研究院合成生物學研究所劉陳立教授及巴黎第七大學的Adrian Daerr博士參與了研究。巴黎第七大學的Agnese I. Curatolo博士,及港大醫學院的周楠博士、趙永峰博士為共同第一作者。
本項研究由法國與香港合作研究計劃(編號:A-HKU712/14)、深圳市孔雀團隊項目和深圳市科技創新委員會基礎研究項目資助。
傳媒查詢
港大生物醫學學院
黃建東教授(電話:3917 6810 | 電郵:jdhuang@hku.hk)
港大生物醫學學院辦公室
陳秀玲 Christina Chan (電話:3917 6334 | 電郵:slcchan@hku.hk)

